STACK service - Python Client Dask
This notebook demonstrates how to use the DEDL Stack Python client to authenticate, manage, and execute parallel, multi-cloud Dask computations on distributed datasets stored across Central Site and LUMI bridge.
Multi-cloud processing with Dask¶
Overview¶
Content¶
DestinE Data Lake (DEDL) Stack Client
Making use of clients context manager
Use Case: Pakistan Flood 2022
Duration: 15 min.¶
DestinE Data Lake utilises a deployment of Dask Gateway on each location (bridge) in the data lake. Dask Gateway provides a secure, multi-tenant server for managing Dask clusters. It allows users to launch and use Dask clusters in a shared, centrally managed cluster environment, without requiring users to have direct access to the underlying cluster backend (e.g. Kubernetes, Hadoop/YARN, HPC Job queues, etc…).
Dask Gateway exposes a REST API to spawn clusters on demand. The overall architecture of Dask Gateway is depicted hereafter.
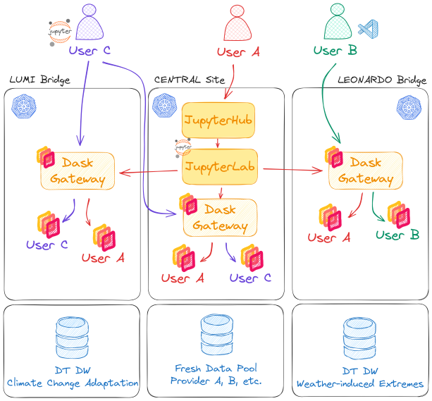
DEDL Dask Gateway¶
Central Site
proxy_address: tcp://dask.central.data.destination-earth.eu:80
LUMI Bridge
proxy_address: tcp://dask.lumi.data.destination-earth.eu:80
Only authenticated access is granted to the DEDL STACK service Dask, therefore a helper class to authenticate a user against the DESP identity management system is implemented. The users password is directly handed over to the request object and is not permanently stored.
In the following, please enter your DESP username and password. Again, the password will only be saved for the duration of this user session and will be remove as soon as the notebook/kernel is closed.
from dask_gateway.auth import GatewayAuth
from getpass import getpass
from destinelab import AuthHandler as DESP_AuthHandler
class DESPAuth(GatewayAuth):
def __init__(self, username: str):
self.auth_handler = DESP_AuthHandler(username, getpass("Please input your DESP password: "))
self.access_token = self.auth_handler.get_token()
def pre_request(self, _):
headers = {"Authorization": "Bearer " + self.access_token}
return headers, Nonefrom rich.prompt import Prompt
myAuth = DESPAuth(username=Prompt.ask(prompt="Username"))DestinE Data Lake (DEDL) Stack Client¶
The DEDL Stack Client is a Python library to facilitate the use of Stack Service Dask. The main objective is to provide an abstraction layer to interact with the various clusters on each DEDL bridge. Computations can be directed to the different Dask clusters by making use of a context manager as given in the following.
from dedl_stack_client.dask import DaskMultiCluster
myDEDLClusters = DaskMultiCluster(auth=myAuth)
myDEDLClusters.new_cluster()myDEDLClusters.get_cluster_url()We can again showcase the execution of standard Python functions on the remote clusters.
In the following we will make use of dask.futures, non-blocking distributed calculations, utilising the map() method for task distribution. Detailed information about dask.futures can be found on the Dask documention.
This approach allows for embarrassingly parallel task scheduling, which is very similar to Function as a Service capabilities.
from time import sleep as wait
def apply_myfunc(x):
wait(1)
return x+1We want to run apply_myfunc() on Central Site and wait for all results to be ready. my_filelist_central represents a filelist to be processed by apply_myfunc().
my_filelist_central = range(20)
with myDEDLClusters.as_current(location="central") as myclient:
central_future = myclient.map(apply_myfunc, my_filelist_central)
results_central = myclient.gather(central_future)results_centralRun computation at LUMI bridge.
my_filelist_lumi = range(32)
with myDEDLClusters.as_current(location="lumi") as myclient:
lumi_future = myclient.map(apply_myfunc, my_filelist_lumi)
results_lumi = myclient.gather(lumi_future)results_lumiLimitations¶
Python libraries use in the local environment need to match, same version, with those available in the Dask Cluster. If this is not the case, you will get a warning, code might work but not guaranteed.
No direct data exchange between Dask Workers across cloud locations possible. Each location acts as atmoic unit, however data can be easily exchanged via storage services such as S3.
Use Case example: Pakistan Flood 2022¶
The complete use case is available on GitHub via https://
The use case demonstrates the multi-cloud capabilities of DEDL following the paradigm of data proximate computing. Data of the Global Flood Monitoring (GFM) service as well as Climate DT outputs, simulated by utilising ERA5 data have been use for flood risk assessment.
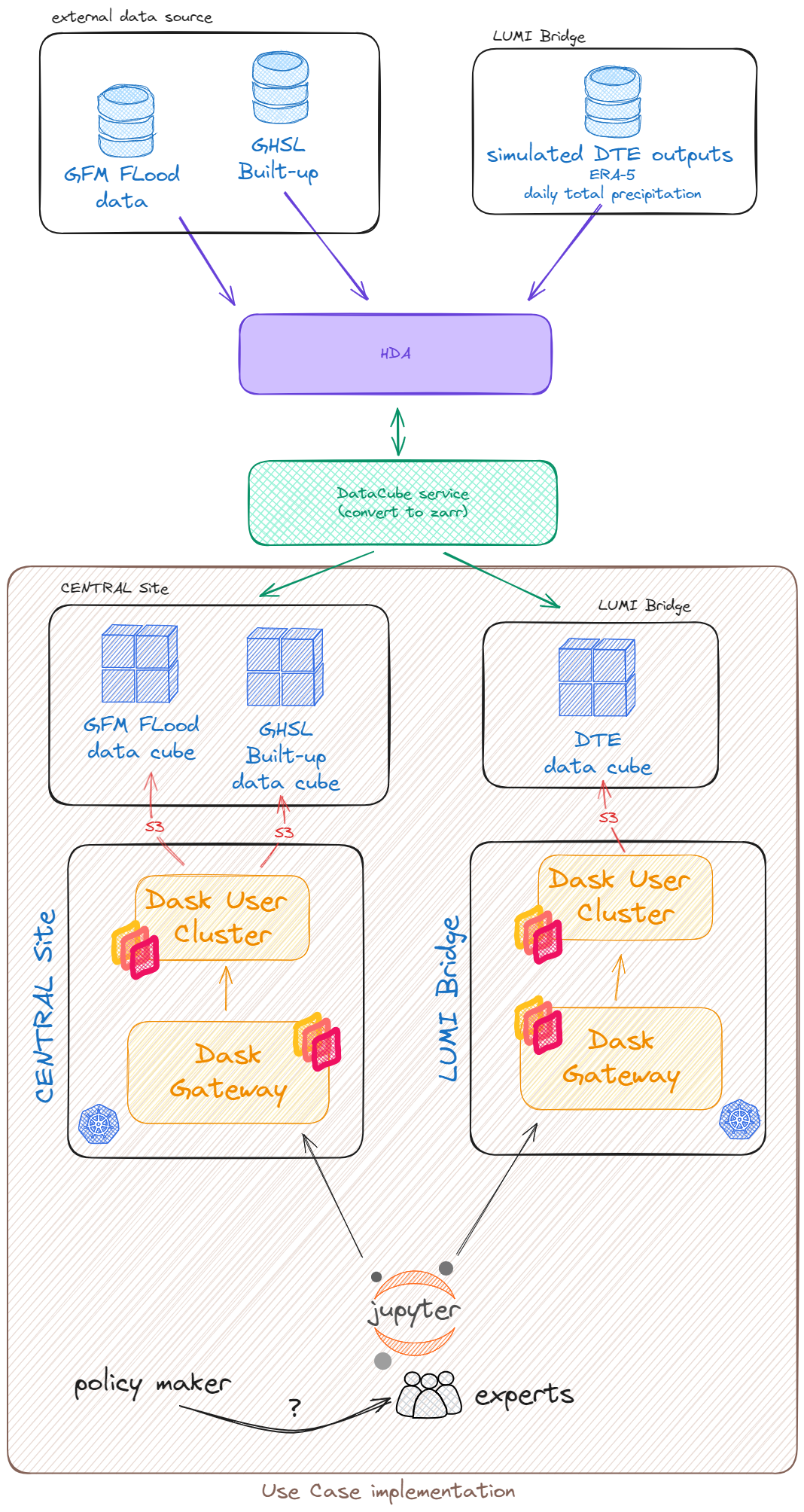
Data is stored as datacubes (zarr format) at Central Site and at LUMI bridge in object storage.
import s3fs
import xarray as xr
xr.set_options(keep_attrs=True)
s3fs_central = s3fs.S3FileSystem(
anon=True,
use_ssl=True,
client_kwargs={"endpoint_url": "https://s3.central.data.destination-earth.eu"})
s3fs_lumi = s3fs.S3FileSystem(
anon=True,
use_ssl=True,
client_kwargs={"endpoint_url": "https://s3.lumi.data.destination-earth.eu"})We can list the data available at Central Site.
s3fs_central.ls("increment1-testdata")Read data stored in S3 bucket at Central Site. The data we want to read is a single Zarr data store representing the GFM flood data over Pakistan for 2022-08-30.
flood_map = xr.open_zarr(store=s3fs.S3Map(root=f"increment1-testdata/2022-08-30.zarr", s3=s3fs_central, check=False),
decode_coords="all",)["flood"].assign_attrs(location="central", resolution=20)
#flood_mapWe now want to run simple computation and compute the flooded area for the this day in August 2022.
flooded_area_ = flood_map.sum()*20*20/1000.
#flooded_area_So far we haven’t computed anything, so lets do the computation now on the Dask cluster.
from rich.console import Console
from rich.prompt import Prompt
console = Console()
flooded_area = myDEDLClusters.compute(flooded_area_, sync=True)
console.print(f"Flooded area: {flooded_area.data} km2")How was that processing routed to Dask Gateway at Central Site?
myDEDLClusters.compute(flooded_area_, sync=True) checks for annotations (attributes) of array and maps that to available Dask Clusters.
Preprocess GFM data at Central Site for visualiation
def preprocess_dataset(data_array: xr.DataArray, method: str):
data_array = data_array.squeeze()
steps = 500 // data_array.attrs["resolution"]
coarsened = data_array.coarsen({'y': steps, 'x': steps}, boundary='trim')
if method == 'median':
data_array = (coarsened.median() > 0).astype('float32')
elif method == 'mean':
data_array = coarsened.mean()
elif method == 'max':
data_array = coarsened.max()
else:
raise NotImplementedError(method)
return data_arrayflood_prep_ = preprocess_dataset(flood_map, 'median')import numpy as np
flood_prep = myDEDLClusters.compute(flood_prep_, sync=True)
flood_prep.rio.write_crs("epsg:4326", inplace=True)
flood_prep = flood_prep.rio.reproject(f"EPSG:3857", nodata=np.nan)Visualise flood data on map.
import leafmap
from attr import dataclass
@dataclass
class Extent:
min_x: float
min_y: float
max_x: float
max_y: float
crs: str
def get_center(self):
return (np.mean([self.min_y, self.max_y]),
np.mean([self.min_x,self.max_x]))
roi_extent = Extent(65, 21, 71, 31, crs='EPSG:4326')
m = leafmap.Map(center=roi_extent.get_center(),
zoom=8, height=600)
m.add_raster(flood_prep, colormap="Blues", layer_name="Flood", nodata=0.)
mRead data stored in S3 bucket at LUMI bridge (Finland). Data we want to read is a datacube generated from ERA-5 representing predicted rainfall data.
rainfall = xr.open_zarr(store=s3fs.S3Map(root=f"increment1-testdata/predicted_rainfall.zarr",
s3=s3fs_lumi,
check=False),
decode_coords="all",)["tp"].assign_attrs(location="lumi", resolution=20)And again run the computation close to the data, therefore at LUMI bridge.
First we compute the accumulated rainfall over Pakistan. Secondly we compute the average rainfall for August 2022 (monthly mean) at global scale.
from datetime import datetime
def accum_rain_predictions(rain_data, startdate, enddate, extent):
rain_ = rain_data.sel(time=slice(startdate, enddate),
latitude=slice(extent.max_y, extent.min_y),
longitude=slice(extent.min_x, extent.max_x))
return rain_.cumsum(dim="time", keep_attrs=True)*1000
# compute accumulated rainfall over Pakistan
acc_rain_ = accum_rain_predictions(rainfall, startdate=datetime(2022, 8, 18),
enddate=datetime(2022, 8, 30),
extent=roi_extent)
acc_rain_ = acc_rain_.rename({"longitude":"x", "latitude":"y"})acc_rain = myDEDLClusters.compute(acc_rain_, sync=True)def acc_rain_reproject(rain):
from rasterio.enums import Resampling
rain.rio.write_nodata(0, inplace=True)
rain.rio.write_crs('EPSG:4326', inplace=True)
return rain.rio.reproject('EPSG:3857', resolution=500, resampling=Resampling.bilinear)
acc_rain = acc_rain_reproject(acc_rain)Visualise forecast data provided by the Digital Twin which could have been used for flood risk assessment or even alerting.
time_dim_len = acc_rain.shape[0]
for day in range(0, time_dim_len):
fpath_str = f"./{day}.tif"
acc_rain[day,:].rio.to_raster(fpath_str,
driver="COG",
overview_count=10)import leafmap
from localtileserver import get_leaflet_tile_layer
m = leafmap.Map(center=roi_extent.get_center(),
zoom=6, height=600)
layer_dict = {}
date_vals = np.datetime_as_string(acc_rain["time"].values, unit='D')
for day in range(0, time_dim_len):
layer_dict[date_vals[day]]= get_leaflet_tile_layer(f"./{day}.tif",
colormap="Blues",
indexes=[1],
nodata=0.,
vmin=acc_rain.min().values,
vmax=acc_rain.max().values,
opacity=0.85)
m.add_local_tile(flood_prep,
colormap="Blues",
nodata=0.)
m.add_time_slider(layer_dict,
layer="Accumluated Rainfall",
time_interval=1.)
mmyDEDLClusters.shutdown()